5 Part 4. Data visualization
5.1 Part 4a. Create a UMAP plot colored by class
Here we can create a UMAP plot of our training data. The UMAP takes MFCCs as features, and each point represents one training sample. In the plot below the images are colored based on class membership.
library(gibbonR)
gibbonID(input.dir="data/BorneoMultiClass",
output.dir="data/BorneoMultiClassthumbnails/",
win.avg='standard',
add.spectrograms=TRUE,
min.freq=400,max.freq=1600,
class='no.clustering')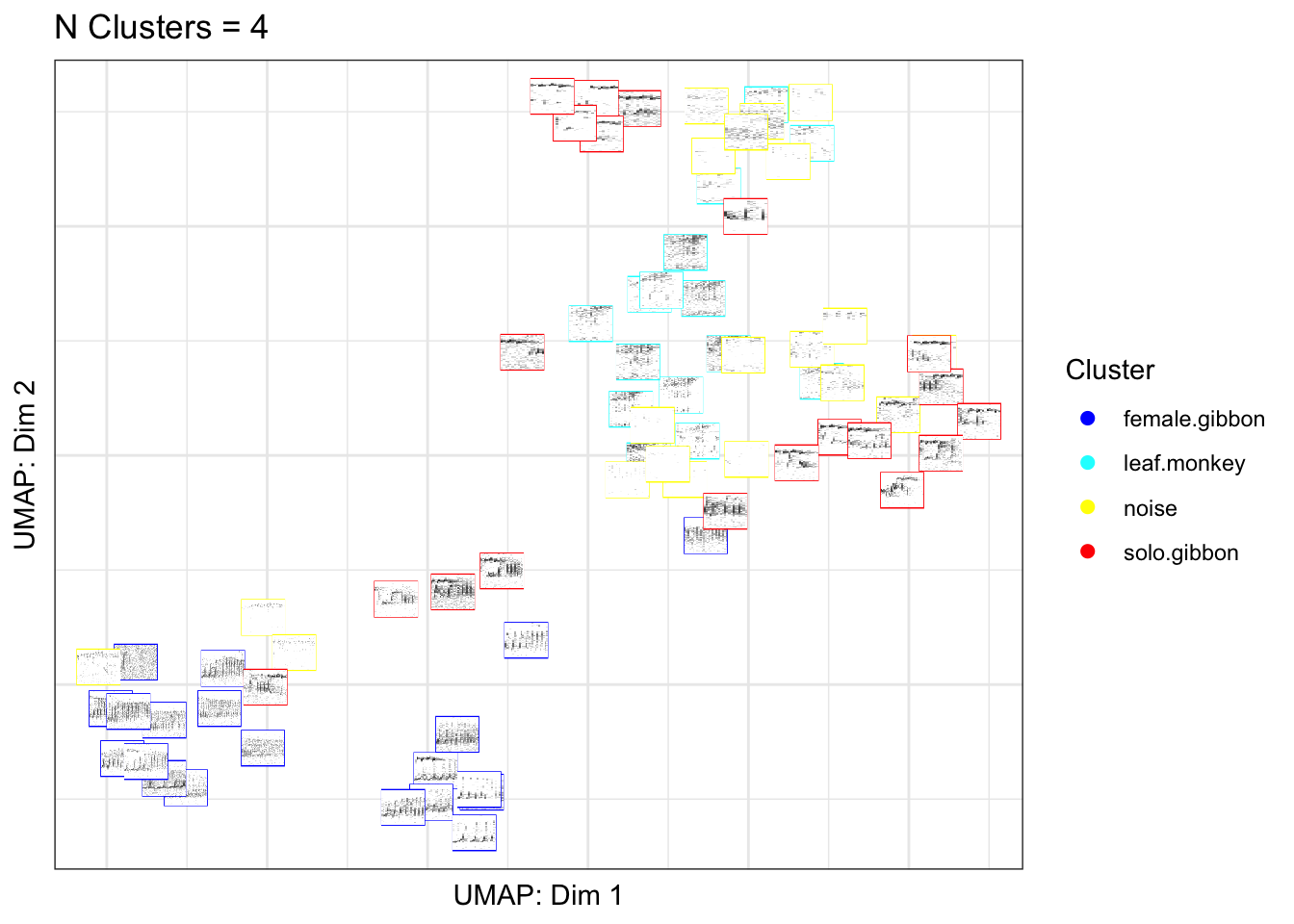
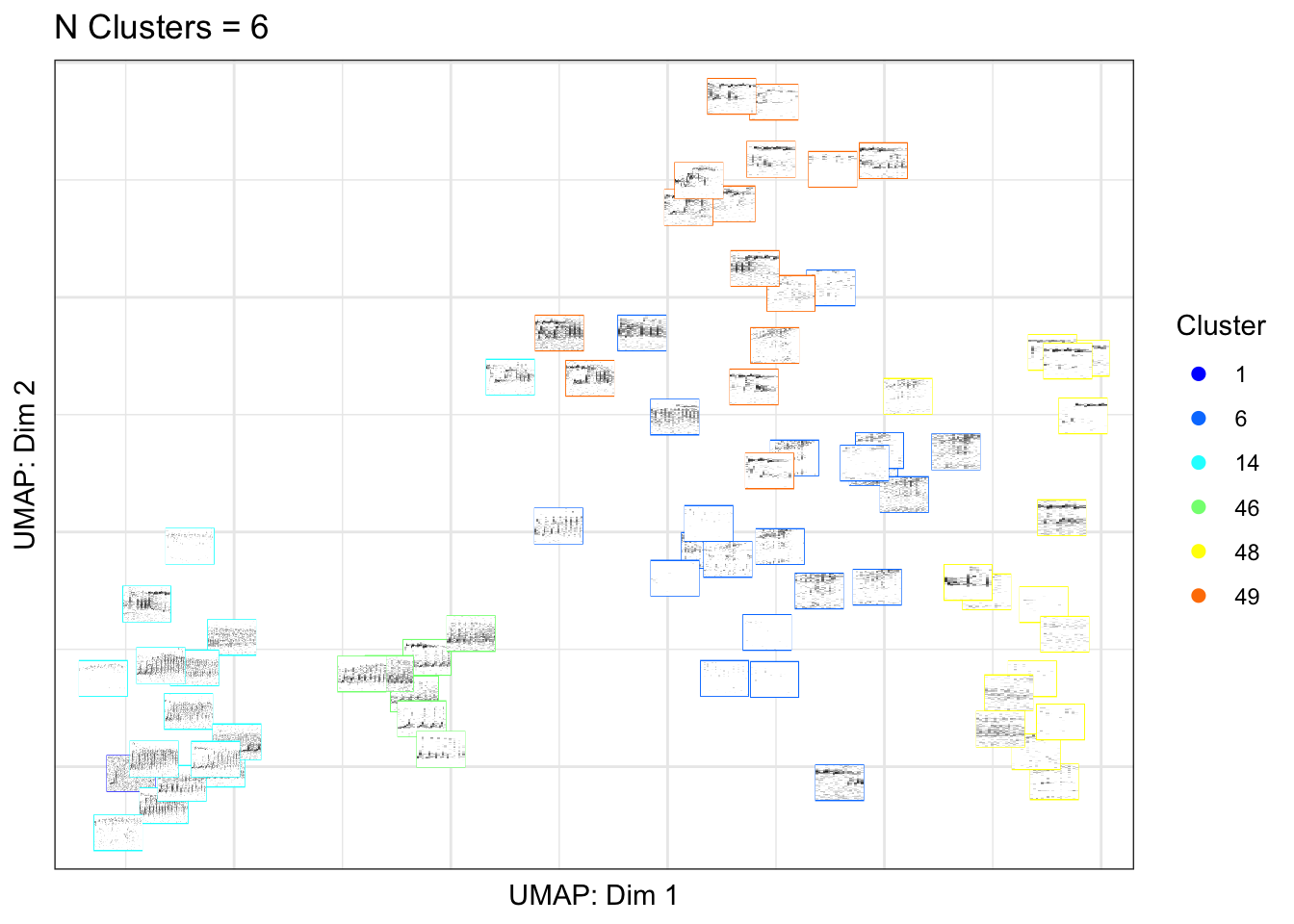
5.2 Part 4b. Create a UMAP plot colored by affinity propagation clustering
In the plot below we use an unsupervised clustering algorithm (affinity propgation clustering) to assign each training sample to a cluster. The spectrogram images below are colored based on cluster assigment.
library(gibbonR)
gibbonID(input.dir="data/BorneoMultiClass",
output.dir="data/BorneoMultiClassthumbnails/",
win.avg='standard',
add.spectrograms=TRUE,
min.freq=400,max.freq=1600,
class='affinity.fixed')